DataFrames Extras#
Some selected functionalities and packages
using DataFrames
using CategoricalArrays
Frequency Tables#
using FreqTables
df = DataFrame(a=rand('a':'d', 1000), b=rand(["x", "y", "z"], 1000))
ft = freqtable(df, :a, :b) ## observe that dimensions are sorted if possible
4×3 Named Matrix{Int64}
a ╲ b │ x y z
──────┼───────────
a │ 85 74 96
b │ 85 93 76
c │ 67 86 75
d │ 86 91 86
you can index the result using numbers or names
ft[1,1], ft['b', "z"]
(85, 76)
getting proportions - 1 means we want to calculate them in rows (first dimension)
prop(ft, margins=1)
4×3 Named Matrix{Float64}
a ╲ b │ x y z
──────┼─────────────────────────────
a │ 0.333333 0.290196 0.376471
b │ 0.334646 0.366142 0.299213
c │ 0.29386 0.377193 0.328947
d │ 0.326996 0.346008 0.326996
and columns are normalized to 1.0 now
prop(ft, margins=2)
4×3 Named Matrix{Float64}
a ╲ b │ x y z
──────┼─────────────────────────────
a │ 0.263158 0.215116 0.288288
b │ 0.263158 0.270349 0.228228
c │ 0.20743 0.25 0.225225
d │ 0.266254 0.264535 0.258258
x = categorical(rand(1:3, 10))
levels!(x, [3, 1, 2, 4]) ## reordering levels and adding an extra level
freqtable(x) ## order is preserved and not-used level is shown
4-element Named Vector{Int64}
Dim1 │
──────┼──
3 │ 0
1 │ 5
2 │ 5
4 │ 0
by default missing values are listed
freqtable([1,1,2,3,missing])
4-element Named Vector{Int64}
Dim1 │
────────┼──
1 │ 2
2 │ 1
3 │ 1
missing │ 1
but we can skip them
freqtable([1,1,2,3,missing], skipmissing=true)
3-element Named Vector{Int64}
Dim1 │
──────┼──
1 │ 2
2 │ 1
3 │ 1
df = DataFrame(a=rand(3:4, 1000), b=rand(5:6, 1000))
ft = freqtable(df, :a, :b) ## now dimensions are numbers
2×2 Named Matrix{Int64}
a ╲ b │ 5 6
──────┼─────────
3 │ 252 243
4 │ 251 254
this is an error - standard array indexing takes precedence
try
ft[3,5]
catch e
show(e)
end
BoundsError([252 243; 251 254], (3, 5))
you have to use Name() wrapper
ft[Name(3), Name(5)]
252
DataFramesMeta.jl#
JuliaData/DataFramesMeta.jl provides a more terse syntax due to the benefits of metaprogramming.
using DataFramesMeta
df = DataFrame(x=1:8, y='a':'h', z=repeat([true,false], outer=4))
| Row | x | y | z |
|---|---|---|---|
| Int64 | Char | Bool | |
| 1 | 1 | a | true |
| 2 | 2 | b | false |
| 3 | 3 | c | true |
| 4 | 4 | d | false |
| 5 | 5 | e | true |
| 6 | 6 | f | false |
| 7 | 7 | g | true |
| 8 | 8 | h | false |
expressions with columns of DataFrame
@with(df, :x + :z)
8-element Vector{Int64}:
2
2
4
4
6
6
8
8
you can define complex operations code blocks
@with df begin
a = :x[:z]
b = :x[.!:z]
:y + [a; b]
end
8-element Vector{Char}:
'b': ASCII/Unicode U+0062 (category Ll: Letter, lowercase)
'e': ASCII/Unicode U+0065 (category Ll: Letter, lowercase)
'h': ASCII/Unicode U+0068 (category Ll: Letter, lowercase)
'k': ASCII/Unicode U+006B (category Ll: Letter, lowercase)
'g': ASCII/Unicode U+0067 (category Ll: Letter, lowercase)
'j': ASCII/Unicode U+006A (category Ll: Letter, lowercase)
'm': ASCII/Unicode U+006D (category Ll: Letter, lowercase)
'p': ASCII/Unicode U+0070 (category Ll: Letter, lowercase)
@with creates hard scope so variables do not leak out
df2 = DataFrame(a = [:a, :b, :c])
@with(df2, :a .== ^(:a)) ## sometimes we want to work on a raw Symbol, ^() escapes it
3-element BitVector:
1
0
0
x_str = "x"
y_str = "y"
df2 = DataFrame(x=1:3, y=4:6, z=7:9)
# $expression inderpolates the expression in-place; in particular this way you can use column names passed as strings
@with(df2, $x_str + $y_str)
3-element Vector{Int64}:
5
7
9
@subset: a very useful macro for filtering
@subset(df, :x .< 4, :z .== true)
| Row | x | y | z |
|---|---|---|---|
| Int64 | Char | Bool | |
| 1 | 1 | a | true |
| 2 | 3 | c | true |
create a new DataFrame based on the old one
@select(df, :x, :y = 2*:x, :z=:y)
| Row | x | y | z |
|---|---|---|---|
| Int64 | Int64 | Char | |
| 1 | 1 | 2 | a |
| 2 | 2 | 4 | b |
| 3 | 3 | 6 | c |
| 4 | 4 | 8 | d |
| 5 | 5 | 10 | e |
| 6 | 6 | 12 | f |
| 7 | 7 | 14 | g |
| 8 | 8 | 16 | h |
create a new DataFrame adding columns based on the old one
@transform(df, :x = 2*:x, :y = :x)
| Row | x | y | z |
|---|---|---|---|
| Int64 | Int64 | Bool | |
| 1 | 2 | 1 | true |
| 2 | 4 | 2 | false |
| 3 | 6 | 3 | true |
| 4 | 8 | 4 | false |
| 5 | 10 | 5 | true |
| 6 | 12 | 6 | false |
| 7 | 14 | 7 | true |
| 8 | 16 | 8 | false |
sorting into a new data frame, less powerful than sort, but lightweight
@orderby(df, :z, -:x)
| Row | x | y | z |
|---|---|---|---|
| Int64 | Char | Bool | |
| 1 | 8 | h | false |
| 2 | 6 | f | false |
| 3 | 4 | d | false |
| 4 | 2 | b | false |
| 5 | 7 | g | true |
| 6 | 5 | e | true |
| 7 | 3 | c | true |
| 8 | 1 | a | true |
Chaining operations#
jkrumbiegel/Chain.jl :Chaining of operations on DataFrame, could be used with DataFramesMeta.jl
using Chain
@chain df begin
@subset(:x .< 5)
@orderby(:z)
@transform(:x² = :x .^ 2)
@select(:z, :x, :x²)
end
| Row | z | x | x² |
|---|---|---|---|
| Bool | Int64 | Int64 | |
| 1 | false | 2 | 4 |
| 2 | false | 4 | 16 |
| 3 | true | 1 | 1 |
| 4 | true | 3 | 9 |
Working on grouped DataFrame#
df = DataFrame(a = 1:12, b = repeat('a':'d', outer=3))
g = groupby(df, :b)
GroupedDataFrame with 4 groups based on key: b
| Row | a | b |
|---|---|---|
| Int64 | Char | |
| 1 | 1 | a |
| 2 | 5 | a |
| 3 | 9 | a |
⋮
| Row | a | b |
|---|---|---|
| Int64 | Char | |
| 1 | 4 | d |
| 2 | 8 | d |
| 3 | 12 | d |
groupby+combine in one line
using Statistics
@by(df, :b, :first = first(:a), :last = last(:a), :mean = mean(:a))
| Row | b | first | last | mean |
|---|---|---|---|---|
| Char | Int64 | Int64 | Float64 | |
| 1 | a | 1 | 9 | 5.0 |
| 2 | b | 2 | 10 | 6.0 |
| 3 | c | 3 | 11 | 7.0 |
| 4 | d | 4 | 12 | 8.0 |
the same as by but on grouped DataFrame
@combine(g, :first = first(:a), :last = last(:a), :mean = mean(:a))
| Row | b | first | last | mean |
|---|---|---|---|---|
| Char | Int64 | Int64 | Float64 | |
| 1 | a | 1 | 9 | 5.0 |
| 2 | b | 2 | 10 | 6.0 |
| 3 | c | 3 | 11 | 7.0 |
| 4 | d | 4 | 12 | 8.0 |
similar in DataFrames.jl - we use auto-generated column names
combine(g, :a .=> [first, last, mean])
| Row | b | a_first | a_last | a_mean |
|---|---|---|---|---|
| Char | Int64 | Int64 | Float64 | |
| 1 | a | 1 | 9 | 5.0 |
| 2 | b | 2 | 10 | 6.0 |
| 3 | c | 3 | 11 | 7.0 |
| 4 | d | 4 | 12 | 8.0 |
perform operations within a group and return ungrouped DataFrame
@transform(g, :center = mean(:a), :centered = :a .- mean(:a))
| Row | a | b | center | centered |
|---|---|---|---|---|
| Int64 | Char | Float64 | Float64 | |
| 1 | 1 | a | 5.0 | -4.0 |
| 2 | 2 | b | 6.0 | -4.0 |
| 3 | 3 | c | 7.0 | -4.0 |
| 4 | 4 | d | 8.0 | -4.0 |
| 5 | 5 | a | 5.0 | 0.0 |
| 6 | 6 | b | 6.0 | 0.0 |
| 7 | 7 | c | 7.0 | 0.0 |
| 8 | 8 | d | 8.0 | 0.0 |
| 9 | 9 | a | 5.0 | 4.0 |
| 10 | 10 | b | 6.0 | 4.0 |
| 11 | 11 | c | 7.0 | 4.0 |
| 12 | 12 | d | 8.0 | 4.0 |
this is defined in DataFrames.jl
DataFrame(g)
| Row | a | b |
|---|---|---|
| Int64 | Char | |
| 1 | 1 | a |
| 2 | 5 | a |
| 3 | 9 | a |
| 4 | 2 | b |
| 5 | 6 | b |
| 6 | 10 | b |
| 7 | 3 | c |
| 8 | 7 | c |
| 9 | 11 | c |
| 10 | 4 | d |
| 11 | 8 | d |
| 12 | 12 | d |
actually this is not the same as DataFrame() as it preserves the original row order
@transform(g)
| Row | a | b |
|---|---|---|
| Int64 | Char | |
| 1 | 1 | a |
| 2 | 2 | b |
| 3 | 3 | c |
| 4 | 4 | d |
| 5 | 5 | a |
| 6 | 6 | b |
| 7 | 7 | c |
| 8 | 8 | d |
| 9 | 9 | a |
| 10 | 10 | b |
| 11 | 11 | c |
| 12 | 12 | d |
Row-wise operations on DataFrame#
df = DataFrame(a = 1:12, b = repeat(1:4, outer=3))
| Row | a | b |
|---|---|---|
| Int64 | Int64 | |
| 1 | 1 | 1 |
| 2 | 2 | 2 |
| 3 | 3 | 3 |
| 4 | 4 | 4 |
| 5 | 5 | 1 |
| 6 | 6 | 2 |
| 7 | 7 | 3 |
| 8 | 8 | 4 |
| 9 | 9 | 1 |
| 10 | 10 | 2 |
| 11 | 11 | 3 |
| 12 | 12 | 4 |
such conditions are often needed but are complex to write
@transform(df, :x = ifelse.((:a .> 6) .& (:b .== 4), "yes", "no"))
| Row | a | b | x |
|---|---|---|---|
| Int64 | Int64 | String | |
| 1 | 1 | 1 | no |
| 2 | 2 | 2 | no |
| 3 | 3 | 3 | no |
| 4 | 4 | 4 | no |
| 5 | 5 | 1 | no |
| 6 | 6 | 2 | no |
| 7 | 7 | 3 | no |
| 8 | 8 | 4 | yes |
| 9 | 9 | 1 | no |
| 10 | 10 | 2 | no |
| 11 | 11 | 3 | no |
| 12 | 12 | 4 | yes |
one option is to use a function that works on a single observation and broadcast it
myfun(a, b) = a > 6 && b == 4 ? "yes" : "no"
@transform(df, :x = myfun.(:a, :b))
| Row | a | b | x |
|---|---|---|---|
| Int64 | Int64 | String | |
| 1 | 1 | 1 | no |
| 2 | 2 | 2 | no |
| 3 | 3 | 3 | no |
| 4 | 4 | 4 | no |
| 5 | 5 | 1 | no |
| 6 | 6 | 2 | no |
| 7 | 7 | 3 | no |
| 8 | 8 | 4 | yes |
| 9 | 9 | 1 | no |
| 10 | 10 | 2 | no |
| 11 | 11 | 3 | no |
| 12 | 12 | 4 | yes |
or you can use @eachrow macro that allows you to process DataFrame rowwise
@eachrow df begin
@newcol :x::Vector{String}
:x = :a > 6 && :b == 4 ? "yes" : "no"
end
| Row | a | b | x |
|---|---|---|---|
| Int64 | Int64 | String | |
| 1 | 1 | 1 | no |
| 2 | 2 | 2 | no |
| 3 | 3 | 3 | no |
| 4 | 4 | 4 | no |
| 5 | 5 | 1 | no |
| 6 | 6 | 2 | no |
| 7 | 7 | 3 | no |
| 8 | 8 | 4 | yes |
| 9 | 9 | 1 | no |
| 10 | 10 | 2 | no |
| 11 | 11 | 3 | no |
| 12 | 12 | 4 | yes |
In DataFrames.jl you would write this as:
transform(df, [:a, :b] => ByRow((a,b) -> ifelse(a > 6 && b == 4, "yes", "no")) => :x)
| Row | a | b | x |
|---|---|---|---|
| Int64 | Int64 | String | |
| 1 | 1 | 1 | no |
| 2 | 2 | 2 | no |
| 3 | 3 | 3 | no |
| 4 | 4 | 4 | no |
| 5 | 5 | 1 | no |
| 6 | 6 | 2 | no |
| 7 | 7 | 3 | no |
| 8 | 8 | 4 | yes |
| 9 | 9 | 1 | no |
| 10 | 10 | 2 | no |
| 11 | 11 | 3 | no |
| 12 | 12 | 4 | yes |
You can also use eachrow from DataFrames to perform the same transformation. However @eachrow will be faster than the operation below.
df2 = copy(df)
df2.x = Vector{String}(undef, nrow(df2))
for row in eachrow(df2)
row[:x] = row[:a] > 6 && row[:b] == 4 ? "yes" : "no"
end
df2
| Row | a | b | x |
|---|---|---|---|
| Int64 | Int64 | String | |
| 1 | 1 | 1 | no |
| 2 | 2 | 2 | no |
| 3 | 3 | 3 | no |
| 4 | 4 | 4 | no |
| 5 | 5 | 1 | no |
| 6 | 6 | 2 | no |
| 7 | 7 | 3 | no |
| 8 | 8 | 4 | yes |
| 9 | 9 | 1 | no |
| 10 | 10 | 2 | no |
| 11 | 11 | 3 | no |
| 12 | 12 | 4 | yes |
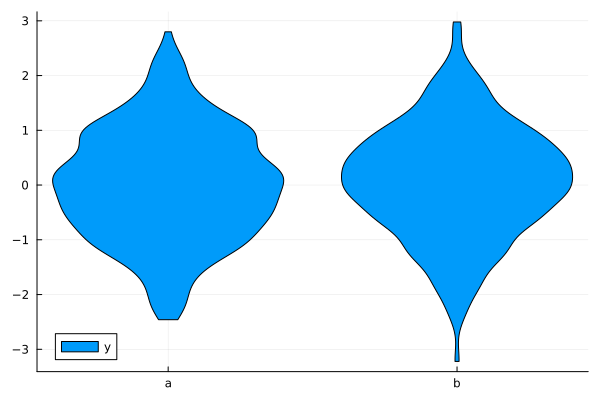
StatsPlots.jl: Visualizing data#
JuliaPlots/StatsPlots.jl you might want to setup Plots package and some plotting backend first
using StatsPlots
A showcase of StatsPlots.jl functions
using Random
Random.seed!(1)
df = DataFrame(x = sort(randn(1000)), y=randn(1000), z = [fill("b", 500); fill("a", 500)]);
a basic plot
@df df plot(:x, :y, legend=:topleft, label="y(x)", seriestype=:scatter)
a density plot
@df df density(:x, label="")
a histogram
@df df histogram(:y, label="y")
a box plot
@df df boxplot(:z, :x, label="x")
a violin plot
@df df violin(:z, :y, label="y")
This notebook was generated using Literate.jl.

