Repressilator#
Repressilator model consists of a biochemical reaction network with three components in a negative feedback loop.
using Catalyst
using ModelingToolkit
using DifferentialEquations
using Plots
repressilator = @reaction_network begin
hillr(P₃, α, K, n), ∅ --> m₁
hillr(P₁, α, K, n), ∅ --> m₂
hillr(P₂, α, K, n), ∅ --> m₃
(δ, γ), m₁ ↔ ∅
(δ, γ), m₂ ↔ ∅
(δ, γ), m₃ ↔ ∅
β, m₁ --> m₁ + P₁
β, m₂ --> m₂ + P₂
β, m₃ --> m₃ + P₃
μ, P₁ --> ∅
μ, P₂ --> ∅
μ, P₃ --> ∅
end
Reactions in the reaction network
reactions(repressilator)
15-element Vector{Catalyst.Reaction}:
Catalyst.hillr(P₃(t), α, K, n), ∅ --> m₁
Catalyst.hillr(P₁(t), α, K, n), ∅ --> m₂
Catalyst.hillr(P₂(t), α, K, n), ∅ --> m₃
δ, m₁ --> ∅
γ, ∅ --> m₁
δ, m₂ --> ∅
γ, ∅ --> m₂
δ, m₃ --> ∅
γ, ∅ --> m₃
β, m₁ --> m₁ + P₁
β, m₂ --> m₂ + P₂
β, m₃ --> m₃ + P₃
μ, P₁ --> ∅
μ, P₂ --> ∅
μ, P₃ --> ∅
State variables in the reaction network
unknowns(repressilator)
6-element Vector{SymbolicUtils.BasicSymbolic{Real}}:
m₁(t)
m₂(t)
m₃(t)
P₁(t)
P₂(t)
P₃(t)
Parameters in the reaction network
parameters(repressilator)
7-element Vector{Any}:
α
K
n
δ
γ
β
μ
Convert to an ODE system#
odesys = convert(ODESystem, repressilator)
To setup parameters (ps) and initial conditions (u₀), you can use Julia symbols to map the values.
p = [:α => 0.5, :K => 40, :n => 2, :δ => log(2) / 120, :γ => 5e-3, :β => 20 * log(2) / 120, :μ => log(2) / 60]
u₀ = [:m₁ => 0.0, :m₂ => 0.0, :m₃ => 0.0, :P₁ => 20.0, :P₂ => 0.0, :P₃ => 0.0]
6-element Vector{Pair{Symbol, Float64}}:
:m₁ => 0.0
:m₂ => 0.0
:m₃ => 0.0
:P₁ => 20.0
:P₂ => 0.0
:P₃ => 0.0
Or you can also use symbols from the ODE system with the @unpack macro (recommended)
@unpack m₁, m₂, m₃, P₁, P₂, P₃, α, K, n, δ, γ, β, μ = repressilator
p = [α => 0.5, K => 40, n => 2, δ => log(2) / 120, γ => 5e-3, β => 20 * log(2) / 120, μ => log(2) / 60]
u₀ = [m₁ => 0.0, m₂ => 0.0, m₃ => 0.0, P₁ => 20.0, P₂ => 0.0, P₃ => 0.0]
6-element Vector{Pair{Symbolics.Num, Float64}}:
m₁(t) => 0.0
m₂(t) => 0.0
m₃(t) => 0.0
P₁(t) => 20.0
P₂(t) => 0.0
P₃(t) => 0.0
Then we can solve this reaction network
tspan = (0.0, 10000.0)
oprob = ODEProblem(repressilator, u₀, tspan, p)
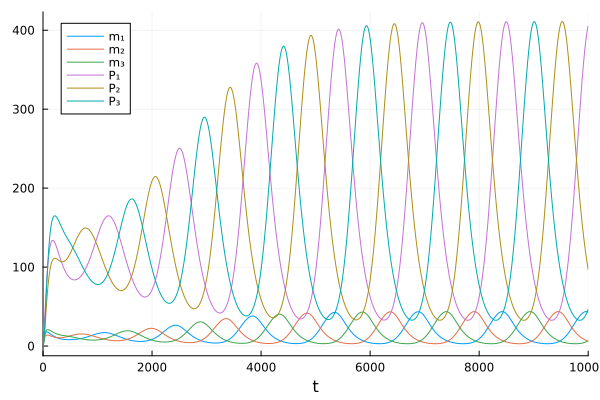
sol = solve(oprob)
plot(sol)
Use extracted symbols.
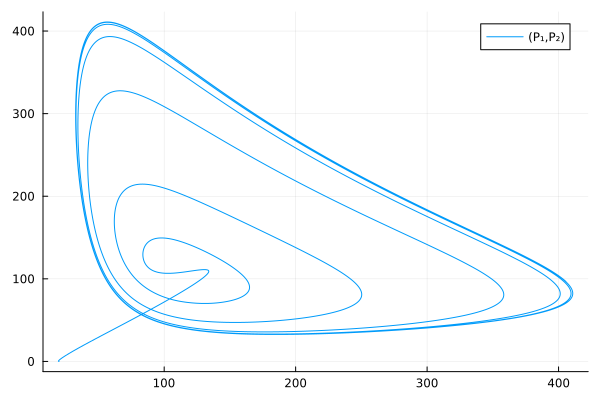
plot(sol, idxs=(P₁, P₂))
Convert to Stochastic Differential Equation (SDE) Models#
Convert the very same reaction network to Chemical Langevin Equation (CLE), adding Brownian motion terms to the state variables.
Build an SDEProblem with the reaction network
tspan = (0.0, 10000.0)
sprob = SDEProblem(repressilator, u₀, tspan, p)
SDEProblem with uType Vector{Float64} and tType Float64. In-place: true
timespan: (0.0, 10000.0)
u0: 6-element Vector{Float64}:
0.0
0.0
0.0
20.0
0.0
0.0
tstops is used to specify enough points that the plot looks well-resolved.
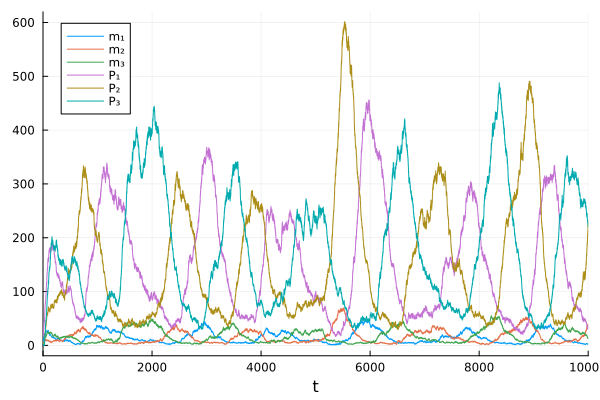
sol = solve(sprob, LambaEulerHeun(), tstops=range(0.0, tspan[2]; length=1001))
plot(sol)
# Using Gillespie's stochastic simulation algorithm (SSA)
Create a Gillespie stochastic simulation model with the same reaction network. The initial conditions should be integers
u0 = [m₁ => 0, m₂ => 0, m₃ => 0, P₁ => 20, P₂ => 0, P₃ => 0]
6-element Vector{Pair{Symbolics.Num, Int64}}:
m₁(t) => 0
m₂(t) => 0
m₃(t) => 0
P₁(t) => 20
P₂(t) => 0
P₃(t) => 0
Create a discrete problem because our species are integer-valued:
dprob = DiscreteProblem(repressilator, u0, tspan, p)
DiscreteProblem with uType Vector{Float64} and tType Float64. In-place: true
timespan: (0.0, 10000.0)
u0: 6-element Vector{Float64}:
0.0
0.0
0.0
20.0
0.0
0.0
Create a JumpProblem, and specify Gillespie’s Direct Method as the solver.
jprob = JumpProblem(repressilator, dprob, Direct(), save_positions=(false, false))
JumpProblem with problem DiscreteProblem with aggregator JumpProcesses.Direct
Number of jumps with discrete aggregation: 3
Number of jumps with continuous aggregation: 0
Number of mass action jumps: 12
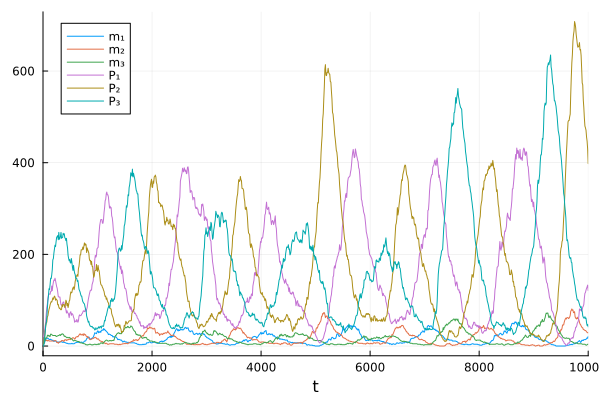
Solve and visualize the problem.
sol = solve(jprob, SSAStepper(), saveat=10.0)
plot(sol)
This notebook was generated using Literate.jl.
